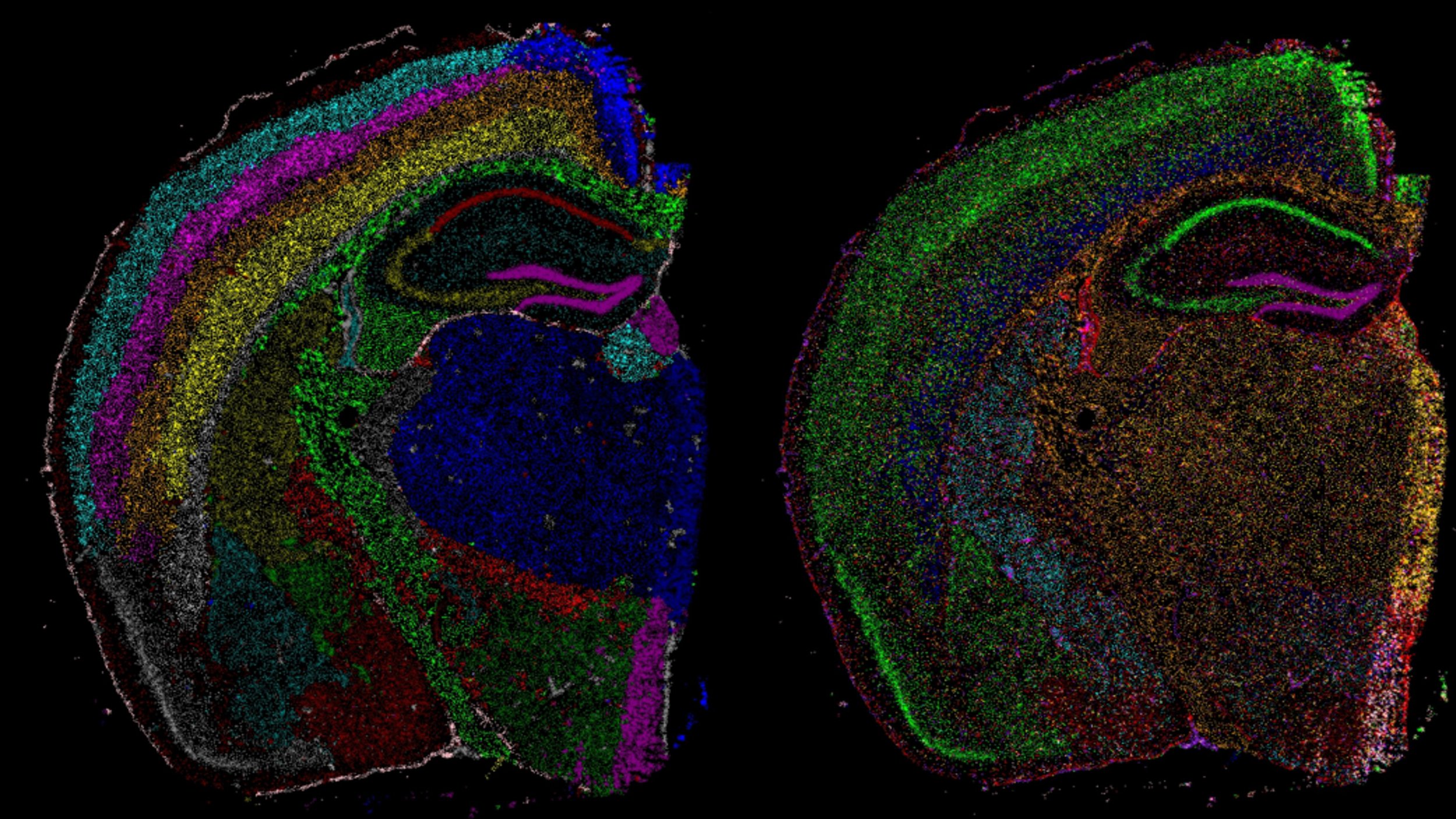
Spatial Biology of animal models
As we identify signature in body fluids that relate to how patients live and perform, we investigate how those omic signatures relate to the tissue alterations present in the affected skeletal muscle tissue.
For this purpose we use spatial technologies on histological sections such as spatial transcriptomics. With the use of this techniques we can then link the biomarkers signatures to changes in tissue morphology (e.g. fibrosis) and understand which cell types are involved.
We focus on several neuromuscular conditions such as Duchenne and Becker Muscular Dystrophies, Limb Girdle Muscular Dystrophies, Facioscapulohumeral Muscular Dystrophy and Inclusion Body Myositis where the cause of the disease is different, but pathophysiological changes are somewhat similar.
This work is typically done in muscle tissue, however given that dystrophin is also expressed in brain we are studying those changes in brain as well by single cell and spatial transcriptomics techniques.
This work is done in collaboration with Dr. Ahmed Mahfouz.
Dystrophin in the brain
Dystrophin is mostly known for its role in striated muscle, however the gene is also expressed in the brain. Several dystrophin isoforms are produced from the DMD gene, some of which are specifically present in the brain and not in muscle such as the Dp427c isoform.
Qirong Mao focuses on mapping and understanding the role of dystrophin in the brain using spatial transcriptomic technologies.
This work is done in collaboration with Dr. Maaike van Putten, Dr. Aurelie Goyenvalle and Dr. Ahmed Mahfouz.
Human Spatial Biology
We use spatial transcriptomics to understand what cells and genes drive tissue changes in neuromuscular conditions.
We are especially interested in the conditions leading to the replacement of skeletal muscle fibers with adipose tissue. To address this questions Laura Heezen analyzed biopsies of patients diagnosed with DMD and BMD, selecting areas where active substitution was visible. With the help of Qirong Mao the studied cell types and genes active at areas of ongoing pathology.
This is one of the different uses of spatial technologies, on which we are currently expanding.
Biomarkers
Our mission is to use biomarkers to understand neuromuscular conditions.
We use a multidisciplinary approach to identify and validate molecular biomarkers in patients’ samples and back up the obtained results with data obtained in animal models. We make us of genomics, transcriptomics, proteomics and metabolomic platforms to identify molecular signatures in body fluids to describe disease progression over longitudinal studies and predict clinically meaningful milestones. This work includes development of quantitative lab methods, statistical modeling and omics data integration.
The work is done in collaboration with Dr. Erik Niks and Dr. Hermien Kan.
RNA Biology
RNA targeting therapies such as exon skipping and read-through stop codons have shown promise in DMD patients. We have shown that dystrophin levels are however reduced compared to healthy counterparts downstream of the mutation site. We further showed that the dystrophin transcript is primarily located in the nucleus by Fluorescent In-Situ Hybridization (FISH). Quantification of nascent transcript has shown that the presence of premature termination codons leads to reduced pre-mRNA synthesis.
To further study the regulation of the synthesized mRNAs, we investigate the RNA binding proteome (interactome) of poly-A transcripts in muscle. We have described the poly-A interactome for proliferating and differentiated muscle cells and we are actively following on this for specific transcripts.
This work is done in collaboration the team of Prof. Annemieke Aartsma-Rus.
Development of statistical methodology
We develop statistical methods tailored for longitudinal omic and clinical data. Our approach is heavily influenced by the specific clinical questions we seek to address, ensuring that our methodologies are not only robust but also directly relevant to the challenges faced in real-world clinical scenarios.
The complexity and multi-dimensionality of this data necessitate sophisticated analytical techniques. We take into account the varying scales, structures, and relationships inherent in multi-omic data, which often includes genomic, proteomic, and metabolomic information, alongside traditional clinical measures. By leveraging innovative statistical frameworks, we aim to extract meaningful insights that can inform patient management and advance scientific understanding in the biomedical field.
Our focus on method development is driven by the need to address specific clinical hypotheses, allowing us to navigate the intricate interplay between biological variables over time. This ensures that the statistical methods we create are not only methodologically sound but also practically applicable, bridging the gap between statistical theory and clinical practice.
Chiara Degan is th statistician in the our group and the work is done in collaboration with Dr. Roula Tsonaka and Prof. Jelle Goeman.