Understanding neuromuscular conditions using biomarkers
Translational Science
Blood Biomarkers in neuromuscular patients
We work on the identification of biomarkers in patients and animal models. We study how molecular signatures in body fluids can predict clinically meaningful milestones. This work includes development of quantitative lab methods, statistical modeling and omics data integration.
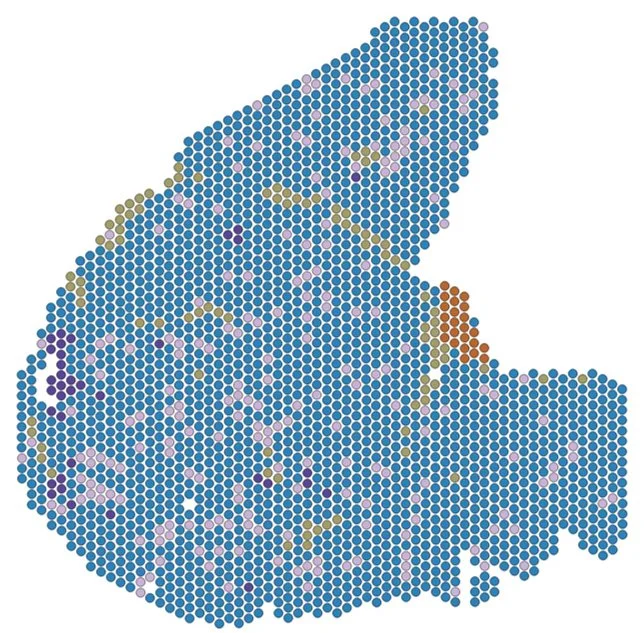
Spatial Biology of patients skeletal muscles
We investigate how omic signatures relate to histological observations in skeletal muscle of patients affected by different neuromuscular conditions. This allows us to link biomarkers to changes in tissue morphology (e.g. muscle fibrosis) and understand which cell types are involved by integrating single cell and spatial data.
Spatial Biology of animal models skeletal muscles
We study the gene expression signature in mouse models to identify genes driving the pathological processes and in response to exposure to therapies.
Basic Science
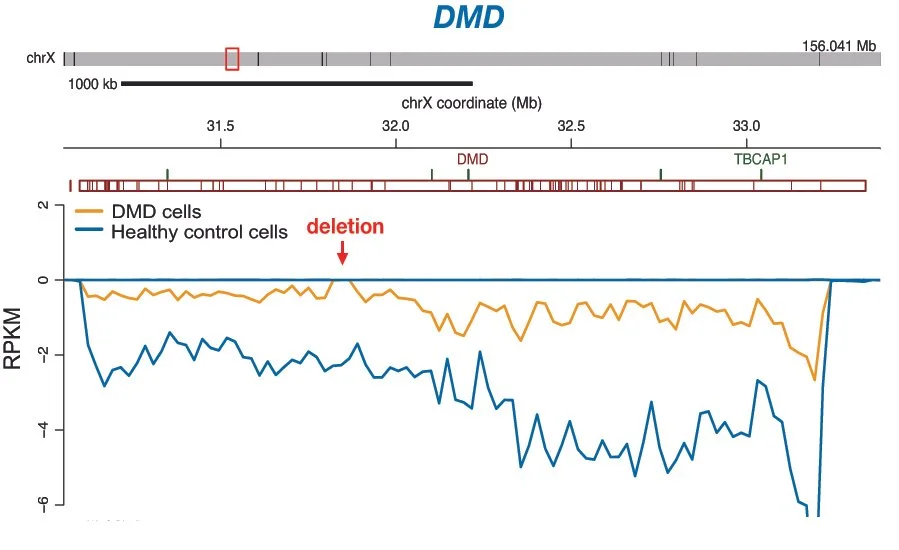
Mapping dystrophin in brain
We use omic data to map dystrophin in brain and to understand the link with behavioural and structural phenotypes observed in dystrophinopathies.
Methods for Statistical Analysis
We develop new statistical tools matching our clinical and biological questions. We focus especially on modeling of biomarkers and clinical data in longitudinal studies with the aim to identify association and predict relevant disease milestones.
RNA Biology
We aim to understand the regulation of neuromuscular genes. By studying transcriptional dynamics and RNA binding proteins we aim to better understand how neuromuscular genes are regulated and improve RNA targeting therapies.
Latest Research
Biomarkers
van de Velde et al, Neurology, 2023
Spatial Biology
Heezen et al, Nat. Commun. 2023
Blood Biomarkers
Ikelaar et al, Nat. Commun., In Press